Front, back, side to side
Now that you have an appreciation of "good" axial EPI time series data we should be able to zip through a review of "good" coronal and sagittal EPIs. This isn't the post to get deep into the reasons why you might want to acquire these prescriptions instead of axial or axial-oblique slices, but here's a short list (and some music) for you to be going on with:
Pros
There are, naturally, drawbacks to coronal and sagittal slices, just as there are for axial slices. I'll mention some of these in more detail below, as we consider the individual artifacts, but here's another brief list:
Cons
Okay then, that's the introduction over with. Let's now put aside the justification for using one prescription over another and look at what constitutes "good" data in the case of coronal and sagittal slices. The features should be immediately recognizable from what you saw in the axial data of the last post.
Parameters
The data we will consider in this post used parameters comparable to those for the axial images in the last post. I used the same single shot, gradient echo EPI sequence on a Siemens Trio/TIM scanner, using the 12-channel head RF coil and a pulse sequence functionally equivalent to the product sequence, ep2d_bold. (See Note 1.) Parameters were: 33 slices, 3 mm slice thickness, 10% slice gap, TR=2000 ms, TE=28 ms, flip angle = 78 deg, 64x64 matrix over a 22.4 cm field-of-view yielding 3.5 mm resolution in-plane, full k-space with phase encoding oriented either left-right (coronal slices) or anterior-posterior (sagittal slices).
To accommodate the scanner's safety software the gradient switching rate had to be reduced slightly compared to the axial prescription, making the echo spacing slightly longer (0.5 ms instead of 0.47 ms) and permitting a maximum of 33 (rather than 34 axial) slices in TR=2000 ms for the coronal and sagittal acquisitions. I acquired just 20 volumes in each time series; enough to demonstrate the temporal features. See Note 2 if you'd like to download the raw data (or the movies and jpeg images) you see below.
Pros
- coronal slices tend to exhibit less dropout of frontal and temporal lobes compared to axial slices.
- coronal slices might permit a smaller field-of-view and higher spatial resolution without signal aliasing than achievable with other prescriptions, assuming your gradient performance and other pulse sequence parameters can be driven sufficiently hard.
- sagittal slices may also show some improved signal in frontal and temporal lobes compared to axial slices, but the real benefit is the unique coverage afforded. You could acquire a single hemisphere, for instance; could be useful in a handful of situations. Alternatively, if you are interested in the whole brain, including cerebellum and perhaps even brain stem, these structures are naturally included in sagittal slices.
- sagittal slices tend to make the most common type of head motion - chin to chest rotations - an in-plane phenomenon which might lead to improved motion correction in post-processing.
There are, naturally, drawbacks to coronal and sagittal slices, just as there are for axial slices. I'll mention some of these in more detail below, as we consider the individual artifacts, but here's another brief list:
Cons
- safety limits on gradient switching (to avoid peripheral nerve stimulation) tend to force the phase encoding direction to be left-right for coronal slices, rendering the EPIs strongly asymmetric. While the absolute level of distortion may actually be very similar to that present in axial slices, the disruption of left-right symmetry can be a shock to your aesthetic sensibility.
- bizarre distortion is also a "feature" of sagittal slices where, as you'll soon see, the distortion can make the frontal lobes look like a duck's bill! But, as before, the absolute level of distortion may not be significantly different to that in axial slices; it's really the unnatural appearance that shocks us. (We ought to be just as outraged at the symmetric distortions in axial slices!)
- perhaps the biggest limitation to both coronal and sagittal prescriptions is the number of slices required to cover the entire brain in the given TR. Slicing along the longest axis of the brain, as done for coronal slices, is clearly the least efficient way to do it. The efficiency of sagittal slices falls somewhere between coronal and axial. And, of course, anything that leads to more (fixed width) slices means that TR might have to get longer. It all depends on your application.
Okay then, that's the introduction over with. Let's now put aside the justification for using one prescription over another and look at what constitutes "good" data in the case of coronal and sagittal slices. The features should be immediately recognizable from what you saw in the axial data of the last post.
Parameters
The data we will consider in this post used parameters comparable to those for the axial images in the last post. I used the same single shot, gradient echo EPI sequence on a Siemens Trio/TIM scanner, using the 12-channel head RF coil and a pulse sequence functionally equivalent to the product sequence, ep2d_bold. (See Note 1.) Parameters were: 33 slices, 3 mm slice thickness, 10% slice gap, TR=2000 ms, TE=28 ms, flip angle = 78 deg, 64x64 matrix over a 22.4 cm field-of-view yielding 3.5 mm resolution in-plane, full k-space with phase encoding oriented either left-right (coronal slices) or anterior-posterior (sagittal slices).
To accommodate the scanner's safety software the gradient switching rate had to be reduced slightly compared to the axial prescription, making the echo spacing slightly longer (0.5 ms instead of 0.47 ms) and permitting a maximum of 33 (rather than 34 axial) slices in TR=2000 ms for the coronal and sagittal acquisitions. I acquired just 20 volumes in each time series; enough to demonstrate the temporal features. See Note 2 if you'd like to download the raw data (or the movies and jpeg images) you see below.
Coronal EPI
Start out by reviewing the cine loop through the twenty frames a few times. Click the 'YouTube' icon on the embedded video to launch an expanded version in a separate tab/window if you prefer (or to download all the videos and jpegs from this post, see Note 2):
Start out by reviewing the cine loop through the twenty frames a few times. Click the 'YouTube' icon on the embedded video to launch an expanded version in a separate tab/window if you prefer (or to download all the videos and jpegs from this post, see Note 2):
The most obvious feature in these coronal images is the left-right distortion which gives the brain a rather perverse, windswept appearance. As I've already mentioned, most of what you're seeing is, in quantitative terms, not appreciably worse than what you deal with in axial slices. Axial slices (with phase encoding set anterior-posterior) do at least maintain a semblance of left-right symmetry, even though we know that many brain regions (particularly frontal lobe) are considerably displaced from their true locations and, indeed, the entire brain appears more oval (stretched) than it should. What disturbs our aesthetic sense in these coronal slices is the disruption by a left-right shearing of what ought to be moderately symmetric anatomy. The problem is especially noticeable for the temporal lobes. (See Note 3.)
If you're wondering why I didn't simply make the phase encoding direction head-foot, the answer is scanner performance and subject comfort. Swapping the read and phase encoding axes means that the rapidly switching read gradient would become left-to-right, from head-to-foot. This might cause more peripheral nerve stimulation, so the scanner's safety software imposes stricter limits on the gradient timing and magnitude parameters in that configuration, and that would translate into longer echo spacing, higher distortion (albeit symmetrically in the brain!) and fewer slices per TR. (See Note 4 for more explanation on stimulus limits.)
Note that throughout the twenty frames of the movie all signal regions, whether strongly distorted (such as temporal lobes) or apparently undistorted (such as the left and right sides of the occipital lobe), remain stable; it is difficult to discern much movement of brain anatomy. The only obvious movement is happening in the neck, because of the pulsation of the carotid arteries (in which blood can be seen flowing as bright vertical lines in a slice in the second row, right-hand side).
What about the ghosts? The N/2 ghosts can be seen in the bottom few rows, but to really see what's going on we want to crank up the background intensity. Here is the same loop of twenty frames but slowed down to 5 frames/sec:
You'll have to take it from me that the ghosts have an intensity about 5% of the brain signal. (You don't really want ghosting more intense than this.) Importantly, the ghosts from the brain signal remain stable over the series.
Let's try to confirm the stability we think we're seeing in the movies by looking at the temporal SNR (TSNR) and standard deviation (SDEV) images:
 |
| TSNR image. (Click to enlarge.) |
 |
| Standard deviation image. (Click to enlarge.) |
The SDEV image reveals the carotid arteries like an angiogram! The superior sagittal sinus is also visible, particularly in the bottom row, as a bright spot atop the midline. In the brain we should expect to see CSF with high SDEV because of cardiac-driven pulsations, just as we saw for the axial slices. And, as before, because gray matter is more metabolically active (and more vascularized) than white matter, GM has higher SDEV than WM. All perfectly normal.
There's an interesting edge that highlights the brain in every slice of the SDEV image. The edge is right along the brain's surface and connects to sulci frequently, strongly suggesting that it arises from pulsating CSF in the meninges, not (whole) head motion or gradient heating. I spent a lot of time inspecting the movies at a host of contrast and zoom combinations, and I couldn't detect by eye a substantial difference for the edges versus the bulk of brain signal. The edges look physiological to me.
Assessing the TSNR image doesn't add much new insight to the picture, except to confirm that there are no brain regions that suffer from a low TSNR when they exhibited high signal in the raw data series. This might happen if, say, there were intense ghosts overlapping portions of the brain that would serve to reduce the TSNR of the overlapped region. (Of course, we would also see these regions having a correspondingly high SDEV, and we don't see any high SDEV ghosts.)
Sagittal EPI
The distortion in the sagittal EPIs makes the brain almost as peculiar as in the coronals. Magnetic susceptibility gradients mutate the frontal lobe into what looks like a peak on a cap. As before, the phase encoding direction is preferentially set anterior-posterior (rather than head-to-foot) to minimize the potential for peripheral nerve stimulation arising from the read gradient train. (See Note 4 again.) Anyway, loop through the movie a few times and see what changes:
Other than the direction of distortion, the features in the movie are quite similar to the previous coronal data and our previous "good" axial data. You now immediately recognize the carotid arteries pulsing away in the neck. If you're wondering why the blood in the carotids appears bright it's because blood is flowing up from the heart where it hasn't experienced the slice-selective RF pulses of prior TR periods. Thus, the "apparent T1" for blood in the carotids is much shorter than the actual T1 for arterial blood. Once the blood has been present in the head for a few seconds, however, it achieves a more conventional T1 steady state, and its signal level is reduced. (The T1 for blood is about 2 seconds at 3 T, so there's only partial relaxation at TR=2 sec.)
Interestingly, in this particular example of sagittal data the subject (me!) seems to be maintaining very good fixation because the eye signal is stable. It is only 40 seconds of data, however, so in a typical fMRI run of a few minutes you should expect to see more eye movements, as we saw previously in the axial data.
There's not much else to talk about, which is a good thing. All signal boundaries appear stationary by inspection. It is difficult to discern with certainty regions of N/2 ghosts so let's follow standard procedure and replay the movie with the background contrasted high:
Now we can clearly identify ghosts - they're most easily identified in the first and last few slices, from the left and right sides of the head, because the volume of brain signal is lower and the background area is larger. These ghosts do not appear by inspection to fluctuate greatly. Most of what's changing is signal in the neck, which one expects from the pulsatile effects of the carotids, perhaps swallowing, small jaw movements and so on. The brain signal appears to be quite stationary.
Okay, time to confirm our cine loop diagnoses with the TSNR and SDEV images:
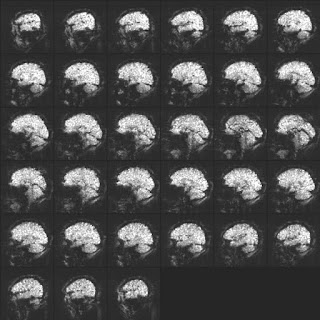 |
| TSNR image. (Click to enlarge.) |
 |
| Standard deviation image. (Click to enlarge.) |
Unsurprisingly, the carotid arteries are a main feature in the SDEV image. Another recognizable feature is the edge enhancement of the brain, much as we saw above in the coronals. Sulci are discernible along the edges again, too, so it's reasonable to assume that this feature arises from pulsatile CSF flow (and for the central few slices perhaps, from the superior sagittal sinus as well). The SDEV image confirms that fluctuations in the N/2 ghosts are low; their standard deviation is barely above that of the background.
The TSNR image shows that in spite of the high degree of distortion the signal in the frontal lobes is viable for fMRI when (as here) the head motion is low. Ugly doesn't imply useless.
Some final thoughts about good data
Now that you have an appreciation of what good time series EPIs look like, and noting that we have only discussed the principal slice prescriptions and a conservative set of parameters in each case, you are ready to start looking at the effects on individual images and time series data when things go wrong; what your physicist will tell you is ‘bad’ or sub-optimal data.
Be warned, though: many of the artifacts in EPI time series have remarkably similar appearance in spite of radically different origins. It can take some time to be able to discriminate between ‘good’ and ‘bad’ data, let alone to become accomplished at discriminating between the different problems when they do arise. That, however, is your task because the better you can diagnose an artifact during your session the faster you can correct it! And with that in mind it's time to shift the focus onto bad data, starting with the next post.
__________________________
Notes:
1. At Berkeley we use a modified version of ep2d_bold, called ep2d_neuro. In these tests there is no functional difference between the two sequences. We have our own local default pulse sequence so that we can have, if desired, thinner slices, a user-defined number of dummy scans, 10 microsec precision in TR, and some other relatively minor tweaks. If you want to replicate these tests then simply set up ep2d_bold with the same parameters as used here.
2. Want the raw data from this post? You can download zip files containing all the DICOM images here:
http://dl.dropbox.com/u/26987499/Good_EPI_Coronal_64x64.zip
http://dl.dropbox.com/u/26987499/Good_EPI_Sagittal_64x64.zip
http://dl.dropbox.com/u/26987499/Good_EPI_Sagittal_64x64.zip
If you don’t already have a DICOM viewer, check out Osirix for Mac OSX (available via a link in the sidebar). ImageJ from NIH - also in the sidebar - has some nice features for ROI analysis, too.
If you want the movies and jpeg images that appear above, these are available here:
http://dl.dropbox.com/u/26987499/Good_EPI_Cor_Sag_jpg_movies.zip
You can use the data, movies and jpegs for any educational purpose you like. No need to acknowledge the source unless you really want to, in which case please cite practiCalfMRI.blogspot.com and the Henry H. Wheeler, Jr. Brain Imaging Center at UC Berkeley.
3. Is the asymmetric L-R distortion a problem for coronal slices? Strictly speaking, it's no worse a problem than it was with axial slices! If you weren't overly concerned with distortion (and its correction) before, with axial slices, then it's a bit feeble to start worrying only when the problem becomes more conspicuous via asymmetry! That said, however, if you apply conventional coregistration algorithms in your analysis pipeline then it's possible that the disruption of left-right symmetry (and in particular the shearing) might cause worse results for coronal than axial slices. I couldn't tell you offhand. Distortion (and its correction) is a complex subject for another post. All I would say at this point is that I wouldn't be dissuaded by distortion alone from exploring a coronal prescription if there were measurable benefits in other domains, such as reduced dropout. I used to use coronal slices for some studies on our old Varian 4 T scanner where it was near impossible to get proper frontal coverage with axial slices. For most of you doing fMRI at 1.5-3 T, though, chances are the reduced coverage along the A-P axis would be the principal reason not to pursue coronal fMRI.
4. The amount of current induced in the subject's (electrically conductive) body is proportional to the cross-sectional area in the plane perpendicular to the switched gradient direction. If the current induced in the body becomes too large it is possible that peripheral nerves will be activated, causing twitching in the subject's muscles.
Given that the dominant gradient is the read (or frequency encode) gradient - the read gradient is generally larger and switched faster than the slice-select and phase encode gradients, as shown in the EPI pulse sequence diagram - then it is this gradient that concerns us the most from a safety perspective. Once the slice selection direction is established it leaves us with just two options for the read gradient direction, the other axis becoming the phase-encoded axis by default.
In understanding the safety limits for switched gradients it is useful to consider the body's three planes as if they act like pick-up coils; loops of wire that can sense changing magnetic fields by having an electric current induced in them. (It's a situation not unlike the RF coils we use in practice to detect the oscillating magnetization! But I digress.) Consider this cartoon showing the effective current loops formed in a subject's body when a gradient is switched in one of three cardinal axes:
So let's consider our options for a coronal EPI slice prescription. Now, the body's cross-sectional area in the plane perpendicular to the H-F axis, i.e. the subject's axial plane, is less than the cross-sectional area in the plane perpendicular to the L-R axis, i.e. the subject's sagittal plane. It's as if the axial plane contains a pick-up loop that's smaller than one in the sagittal plane, as illustrated by the black loops in the cartoon. The induced currents in the subject will be lower if we choose H-F for the read gradient direction assuming, as is virtually always the case for conventional fMRI, that the read gradient is larger than the phase encoding gradient. Thus, the phase encoding axis becomes L-R.
Suppose that you don't particularly like the asymmetric distortion of your coronal EPIs, and you decide to try and swap the read and phase encode gradient directions. What are the consequences? The good news is that you do indeed return the L-R axis of the EPIs to being distortion-free, and any stretches/compressions would now be H-F. But there's a price: we run a higher risk of peripheral nerve stimulation. Accordingly, the scanner's software reduces the gradient timing and amplitude parameters (i.e. the hardware performance is reduced) to maintain a safe situation for the subject, thereby reducing our ability to achieve a given spatial resolution and brain coverage (via increased minimum TE, for example) than were achievable in the previous situation, with the read axis H-F. What's more, by slowing down the gradients the minimum echo spacing actually increases, making the absolute level of distortion higher with the phase encoding H-F than it was when it was L-R. So much for symmetry!
That's coronal slices dealt with. A quick glance at the cartoon reveals the preferred read gradient axes for the other two cardinal slice prescriptions. For axial slices, the preferred read gradient direction is L-R, making the phase encode axis A-P. For sagittal slices the preferred read gradient direction is H-F, making the phase encode axis A-P as well.
Another note on the subject of peripheral nerve stimulation: note that the effective current loops we've considered assume that the subject has his hands by his side and his legs uncrossed. Linking hands/feet will increase the size of the effective current loop, especially in the coronal plane where the current loop was already largest! Crossed arms and/or legs may mean that the effects of switching the slice select or phase encode gradients - gradients that we ignored in determining which axis to use for the read gradient - may start to induce peripheral nerve stimulation in the subject. The scanner software assumes that you're not creating artificially large current paths! (I've done these tests myself, as it happens. We have a perfusion sequence with an aggressive EPI readout that is just about able to twitch the trapezius muscles in my back. But if I clasp my hands together I can get reliable twitching, no problem at all! The twitching stops the moment I release my hands.)
What's the bottom line, here? Well, other than the implications for (a)symmetric distortion that depends on the slice prescription, you should now be able to understand how a set of parameters that can be used to acquire EPIs in one orientation may be precluded (by the scanner) for another. What might have seemed a trivial step of attempting to swap the the read and phase encode directions actually has safety implications. Your subject sure appreciates it, even if you don't!

No comments:
Post a Comment