In an earlier post I presented three starting protocols for the CMRR version of SMS-EPI, referred to as the MB-EPI sequence here. I'll use italics to indicate a specific pulse sequence whereas SMS-EPI, no italics, refers to the family of simultaneous multi-slice methods. In this post I'll develop a similar set of three starting protocols for the Massachusetts General Hospital (MGH) version of SMS-EPI, called Blipped-CAIPI. I'm going to build upon the explanations of the last post so please cross reference for parameter explanations and background.
As for the previous post there are several things to bear in mind. This series is Siemens-centric, specifically Trio-centric. While many of the concepts and parameter options may apply to other platforms there will be minor differences in parameter naming conventions and, perhaps, major differences in implementation that you will need to consider before you proceed. For Siemens users, I am running aging software, syngoMR version B17A. The age of the software and the old reconstruction board on the scanner means that you can expect to see much faster reconstruction on a newer system. I hope, but cannot guarantee, that the actual image quality and artifact level won't differ massively from a Trio running VB17A to a new Prisma running VE11C. I'll keep you updated as I learn more.
Preliminaries
As before, for this post I am going to be using a 32-channel receive-only head coil. The SMS-EPI sequences can be made to work with a 12-channel coil but only in a reduced fashion because the 12-channel coil has minimal receive field heterogeneity along the magnet z axis - the struts run parallel with the magnet axis except at the coil's rear, where they converge - and generally we want to do axial slices (along z) for fMRI. I don't yet know whether SMS-EPI would work well on the 20-channel head/neck coil on a Prisma, it's something I hope to investigate in the near future. But a 64-channel head/neck coil on a Prisma will definitely work for SMS-EPI. Better or worse than a 32-channel coil on a Prisma? I have no idea yet.
The Blipped-CAIPI sequence version 2.2 was obtained through a C2P (Core Competence Partnership) with MGH. Installation was a breeze: a single executable to port to the scanner and one click, done. The development team offers an informative but brief 7-page manual which will be useful to anyone who has read the SMS-EPI literature and has a basic understanding of how SMS works. It's not a starting point for everyday neuroscience, however. The manual mentions a .edx (protocol) file as a starting point for 2, 2.5 and 3 mm resolution scans, but in the file I downloaded for VB17A the contents didn't include it. Perhaps contact MGH if you are on another software version and you'd like a .edx file rather than building your own protocol, e.g. by recreating what you see here.
General usage issues
My immediate sense on initial testing was that overall performance of Blipped-CAIPI is quite similar to that of MB-EPI. Timing parameters and spatial resolution limits come out to be very close because they are primarily functions of the scanner hardware rather than the pulse sequence implementation. The small differences arise out of the implementation, the most significant of which is the default RF pulse type and duration. With MB-EPI one sets the duration explicitly, subject to the constraints of the RF amplifier peak output, slice thickness, cross-talk and SAR. With Blipped-CAIPI the RF duration is controlled indirectly, via a VERSE factor. More on that parameter below.
As for MB-EPI, I recommend enabling the Prescan Normalize option and instruct the scanner to save both the normalized and the raw images. The reconstruction overhead is minuscule, while a factor of two more data is a small price to pay for flexibility. However, you may prefer to acquire and use your own receive field correction map in post processing, as the Human Connectome Project did. I don't have specific recommendations today on how best to acquire your own receive field maps, but it's on the list of topics to be covered in this post series on SMS methods.
Both the Blipped-CAIPI and MB-EPI sequences use what is called in the literature the "slice-GRAPPA" reconstruction method (Setsompop et al. 2012). The "split-slice-GRAPPA" recon (Cauley et al. 2014) can be used instead by enabling the LeakBlock parameter on the Special tab of the MB-EPI sequence. In Blipped-CAIPI, however, I think the split-slice-GRAPPA is enabled automatically if one turns on conventional GRAPPA for in-plane acceleration. It's also my understanding that split-slice-GRAPPA (a.k.a. LeakBlock) should be used only when using in-plane GRAPPA with SMS. Still with me? I apologize on behalf of MRI physicists everywhere for a dog's breakfast of nomenclature. Anyway, I'll try to get more information for a recon-specific post at a later date. I'm still feeling this stuff out for myself.
Blipped-CAIPI parameter options
Single band reference images: (Called SBref for MB-EPI.) The single band reference images option is apparently enabled on the Special card, but it is inaccessible and the single band reference images aren't actually saved on my VB17A version. I assume this is just a bug.
Dummy scans: A feature in Blipped-CAIPI is the ability to set dummy scans for the single band reference data independent of separate dummy scans inserted between the reference data and the accelerated time series (the latter is what you think of as "your data"). This assures proper T1 steady states for both the reference and time series data (in the absence of motion) even though the reference data are acquired as quickly as possible (minimum TR). With MB-EPI one can simply insert a handful of volumes of no interest and ignore these in the final time series, so there's minimal practical difference.
VERSE factor: I have no experience with VERSE so I'm going to simply quote from the MGH manual:
Apply the Variable-rate selective excitation (VERSE) method12 to reduce peak voltage and SAR of the MultiBand pulse. When set to level 1, the VERSE method is not used and a standard [Shinnar-LeRoux] SLR pulse is played. The higher the VERSE value, the more reduction in peak voltage and SAR. This comes at a cost of slice profile distortion at off-resonance. The sequence only allows VERSE factors at which profile distortions are generally in an acceptable range. However, one should still set the VERSE level to the lowest level that will not cause RF clipping or exceed SAR limits.
12 Conolly S, Nishimura D, Macovski A, Glover, G. (1988). Variable rate selective excitation. Journal of Magnetic Resonance 78: 440–458.
Variable Rate Excitation (VERSE)
The true RF power is determined by the power integral of the RF over time. By varying the slice selection gradient with time, the k-space representation of the RF pulse is no longer mapped linearly to the time domain, and power can be reduced without modifying the slice profile. This is the principle of VERSE (30). Power reduction is achieved by slowing down k-space traversal at coordinates where most energy needs to be deposited (i.e., the peak of the [multiband] RF waveform) by temporarily reducing the amplitude of the slice selection gradient. Time lost by doing this can be recovered by speeding up at times of low RF amplitude. VERSE has great potential to reduce the power of a pulse but is very sensitive to off-resonance effects (30). Whereas a standard RF pulse off-resonance only experiences a slice shift, the sensitivity of a VERSE pulse to off-resonance varies with time due to the varying ratio between the gradient strength and the magnitude of the local field inhomogeneity, which can lead to a corrupted slice profile as demonstrated in the original study (30). In practice, the low extent to which VERSE needs to be applied to 180 refocusing pulses with moderate multiband factors (MB < 4) at 3 Tesla (T), still allows an acceptable slice profile and high effective bandwidth-time product (31).
So, we should expect to see enhanced signal dropout in the usual problem regions - frontal and temporal lobes, deep brain regions - if we enable the VERSE method. In all the tests I show below I left the VERSE factor at unity, i.e. disabled. I never ran into an RF clipping or SAR-limited situation for flip angles of 45 degrees. Perhaps I'll look at the use of VERSE as a sub-component of a future post on slice-to-slice cross-talk. Until then I'll leave VERSE turned off.
SMS shift: This is the blipped-CAIPI field-of-view (FOV) shift factor from which the pulse sequence derives its name (Setsompop et al. 2012). Here I set the SMS shift to 3 (i.e. FOV/3) based on hints from MGH and the literature. I haven't explored the consequences of different shifts. The MB-EPI sequence sets the FOV shift automatically, but the default without in-plane acceleration appears to be FOV/3.
Number of slices: The Blipped-CAIPI sequence forces you to use an odd number of slice packets, where the number of slice packets is the total number of slices divided by the SMS factor. For example, it won't allow 60 slices when the SMS factor is 6, but 54 or 66 slices are allowed. The restriction against even multiples of the SMS factor is to reduce the potential slice-to-slice cross-talk, an issue I'll go into in depth in the next post because we also need to consider motion when considering cross-talk. (In the mean time, see Note 1.) All I can say right now is that I've not seen much cross-talk (on a stationary phantom) when using even multiples of the SMS factor with MB-EPI.
Compression factor: This is a way to reduce the total number of effective channels in the RF coil through software combination. One gives up unique spatial information in exchange for less computation and faster reconstruction. Here's the description from the MGH manual:
Compression Factor: The amount of compression applied in a Geometric Coil Compression (GCC) scheme to reduce the number of effective coil channels and hence speed up the slice-GRAPPA reconstruction. The Compression Factor can be set to within the range of 1-4. At level 1, no compression is applied. At level 2, coil channels are compressed down by 50% (e.g. a 32-channel coil is compressed down to 16 virtual channels). At level 4, coils are compressed down to 25% (e.g. a 32-channel coil is compressed down to 8 virtual channels). Compression should mainly be used for high channel count arrays such as a 32 or 64 channel coil, where the slice-GRAPPA computation is more intensive and reconstruction time is longer. At level 2 compression (for these high channel count cases), no degradation in reconstruction performance is observed while achieving a fast image reconstruction (typically real-time in most cases). At compression level 4, some minor degradation in reconstruction performance can be observed for high SMS factors acquisitions.
Kernel size: This is another parameter that is set automatically in the MB-EPI sequence, but with Blipped-CAIPI we have three options: 3x3, 5x3 and 5x5. This from the MGH manual:
Kernel Size: Size of the sliceGRAPPA kernel (ky × kx) used to reconstruct the images. Larger kernel sizes typically provide better reconstruction with the cost of longer reconstruction time. For low SMS factors (2-4), a 3×3 kernel size is sufficient. At higher SMS factors, a larger kernel size should be used.
I had no idea how the kernel size would affect image quality so this became the subject of preliminary testing, along with the Compression Factor as a way to influence reconstruction speed.
Blipped-CAIPI reconstruction speed
The reconstruction speed can be a major factor when deciding whether to use SMS or not. While the actual reconstruction performance depends on your software and recon hardware and mine are both rather old (VB17A with a Step IV MRIR), I thought it might be useful to give you a sense of the relative performance. I ran three throwaway tests with the following parameters fixed: 100-volume time series, 2 mm isotropic voxels, SMS factor of 6, FOV/3 CAIPI shift, TR=1000 ms, 66 total slices. I was particularly interested to see how much the kernel size and the compression factor (CF) affected reconstruction speed.
3x3 kernel, CF = 1
The first image appeared after 40 sec, and each subsequent volume took 2.2 sec to process so that the final image reconstructed 4 min 20 sec after acquisition start.
5x5 kernel, CF = 1
The first image appeared at 3 min 15 sec, long after the acquisition of all 100 images had finished. The volumes then reconstructed at a rate of ~2.6 sec/volume so that the final image reconstructed 7 min 30 after acquisition start.
5x5 kernel, CF = 2
The first image appeared after 50 sec, and each subsequent volume took ~2.2 sec to process so that the the final image appeared 4 min 25 sec after acquisition start.
The reconstruction is slow whichever way you look at it. But increasing the kernel size without also using CF > 1 makes the recon very slow indeed. Given the radical effects of CF and kernel size on reconstruction time, what can we say about image quality? Does speed come at a price we can afford to pay? Let's take a look at some specific acquisition parameter sets and then look at the effects of reconstruction parameters on image quality.
Blipped-CAIPI starting point protocols
Here are three starting protocols designed to match as closely as possible the parameters given in the previous post, for the MB-EPI sequence:
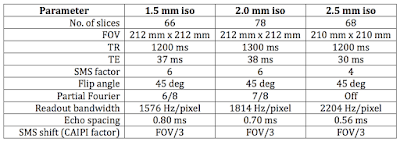 |
| Main parameters for three starting Blipped-CAIPI protocols. (Click to enlarge.) |
The full parameters are available via Dropbox in this PDF. As for the previous MB-EPI starting protocols, the number of slices for 1.5 mm isotropic resolution is insufficient to cover the whole brain. Expect to increase the TR and number of slices by about 50% for whole brain. Alternatively, you might consider increasing the SMS factor up to 8 to improve the coverage, but I haven't tested it and I would be concerned at the potential leakage artifacts that might result. (See Note 2.)
The coverage for the 2.5 mm isotropic voxel protocol is more than sufficient for whole brain when using 68 slices in a TR of 1200 ms. But if you specifically want shorter TR, down to 800 ms or so, you might consider setting the MB factor to 6 rather than 4.
The in-plane parameters are very nearly identical between Blipped-CAIPI and MB-EPI, which is to be expected given that the SMS scheme applies in the slice dimension. Small differences arise out of the duration of the SMS excitation pulse for each pulse sequence, and these lead to slight differences in minimum TE in particular. Any differences in read gradient bandwidth and echo spacing are negligible.
As for MB-EPI, the 2 mm isotropic voxel protocol for Blipped-CAIPI uses an echo spacing (of 0.70 ms) that is in the middle of the range 0.6-0.8 ms where mechanical resonances for axial slices are expected to be worst on a Trio. There is insufficient gradient strength to get below 0.6 ms, while setting the echo spacing at 0.8 ms necessitates a TE greater than 40 ms. This could be offset by setting 6/8ths partial Fourier to get back below 40 ms, but that is likely trading one potential artifact (slightly higher ghosting) for another (slightly higher dropout). In my phantom tests the ghosting from the echo spacing of 0.70 ms weren't terrible (see the example data below). But you should verify on your scanner that such an echo spacing doesn't produce show-stopping ghosts because the mechanical resonances differ from installation to installation.
Blipped-CAIPI performance on a phantom
My first task was to determine a suitable non-SMS control acquisition. I could have set up the product ep2d_bold sequence, say, with all the parameters matched to the SMS protocols except the TR, but I wanted to do a quick comparison between MB-EPI and Blipped-CAIPI and so I'm using the single band reference (SBref) images available from the MB-EPI acquisition as the standard to match. Some things to note. Firstly, the contrast in the SBref images may well be different to SMS because the TR is different. In a reasonably homogeneous gel phantom with a few air bubbles this isn't likely to be a big deal, but it does mean we can expect some intensity differences. Secondly, I want to emphasize that this is a throwaway comparison. Let me state for the record that I don't have a preference, all I have is more experience with MB-EPI than with Blipped-CAIPI. You may interpret any differences you see below any way you like, but I would caution against over interpreting anything you see. For starters I didn't even try to reproduce what I've done and you know that's a bad habit!
The following tests were conducted on an FBIRN gel phantom and with the 32-channel head coil. Each figure comprises four panels to show:
- Top left = SBref "reference" images
- Top right = SMS images acquired with MB-EPI
- Bottom left = SMS images acquired with Blipped-CAIPI, processed with a 5x5 recon kernel and CF = 1
- Bottom right = SMS images acquired with Blipped-CAIPI, processed with a 5x5 recon kernel and CF = 2
Let's begin by looking in the image plane, that is, in the conventional Siemens "mosaic" display. I'll use two intensity settings, the first to highlight any artifacts visible on the phantom and the second with the background scaled to reveal the N/2 ghosts and residual aliasing/leakage from the SMS reconstructions. Please note that the different number of slices in the different sequences means that there isn't necessarily a direct correspondence between the panels showing MB-EPI and Blipped-CAIPI data. The bubbles and cracks in the gel are your "anatomical references" to determine which slice is which.
There are no obvious differences visible on the phantom for 2.5 mm isotropic resolution with SMS factor 4:
 |
| Signal contrasted 2.5 mm isotropic resolution scans, SMS=4: TL: SBref. TR: MB-EPI. BL: Blipped-CAIPI, 5x5 kernel, CF=1. BR: Blipped-CAIPI, 5x5 kernel, CF=2. (Click to enlarge.) |
Residual aliasing is less prominent than the N/2 ghosts, although the ghosts are sharper for Blipped-CAIPI and there is a suspicion of residual aliasing in the MB-EPI artifacts:
 |
| Ghost contrasted 2.5 mm isotropic resolution scans, SMS=4: TL: SBref. TR: MB-EPI. BL: Blipped-CAIPI, 5x5 kernel, CF=1. BR: Blipped-CAIPI, 5x5 kernel, CF=2. (Click to enlarge.) |
I don't see any obvious signal level differences for 2 mm isotropic resolution and SMS factor 6:
 |
| Signal contrasted 2 mm isotropic resolution scans, SMS=6: TL: SBref. TR: MB-EPI. BL: Blipped-CAIPI, 5x5 kernel, CF=1. BR: Blipped-CAIPI, 5x5 kernel, CF=2. (Click to enlarge.) |
But there are now clearly differences in the ghosts. We can use the SBref images' ghosts as our standard to recognize that the Blipped-CAIPI ghosts are crisper as well as more intense, while the MB-EPI ghosts are diffuse and exhibit multiple structures consistent with residual aliasing:
 |
| Ghost contrasted 2 mm isotropic resolution scans, SMS=6: TL: SBref. TR: MB-EPI. BL: Blipped-CAIPI, 5x5 kernel, CF=1. BR: Blipped-CAIPI, 5x5 kernel, CF=2. (Click to enlarge.) |
We seem to have a choice between more intense ghosts or more intense residual aliasing. So what happens when we push to 1.5 mm isotropic resolution and SMS factor of 6? We now see that there are artifacts visible on the phantom. I've picked a few out using orange arrows for the bright artifacts blue arrows for the dark artifacts:
 |
| Signal contrasted 1.5 mm isotropic resolution scans, SMS=6: TL: SBref. TR: MB-EPI. BL: Blipped-CAIPI, 5x5 kernel, CF=1. BR: Blipped-CAIPI, 5x5 kernel, CF=2. (Click to enlarge.) |
At the ghost level we again see better delineated N/2 ghosts for Blipped-CAIPI and diffuse ghosts for MB-EPI, but the artifacts clearly extend across the entire (signal-free) background in both. Scaling the background up to see the artifacts causes the background for the SBref images to saturate, indicating that the artifacts in the accelerated images are now a lot higher than for SBref, and a lot higher than for the prior 2.5 mm or 2 mm protocols:
 |
| Ghost contrasted 1.5 mm isotropic resolution scans, SMS=6: TL: SBref. TR: MB-EPI. BL: Blipped-CAIPI, 5x5 kernel, CF=1. BR: Blipped-CAIPI, 5x5 kernel, CF=2. (Click to enlarge.) |
Now let's look in the slice dimension (reconstructed from the matrix of slices). This confirms no obvious artifacts for 2.5 mm isotropic resolution, SMS factor 4:
 | ||
| Through plane reconstruction for 2.5 mm isotropic resolution scans, SMS=4: TL: SBref. TR: MB-EPI. BL: Blipped-CAIPI, 5x5 kernel, CF=1. BR: Blipped-CAIPI, 5x5 kernel, CF=2. (Click to enlarge.) |
Nothing obvious for the 2 mm isotropic resolution, SMS factor 6:
 |
| Through plane reconstruction for 2 mm isotropic resolution scans, SMS=6: TL: SBref. TR: MB-EPI. BL: Blipped-CAIPI, 5x5 kernel, CF=1. BR: Blipped-CAIPI, 5x5 kernel, CF=2. (Click to enlarge.) |
But the artifacts again become apparent for 1.5 mm isotropic resolution, SMS factor 6. The orange arrow indicates a dark band on the MB-EPI image, green arrows indicate bright horizontal bands in the Blipped-CAIPI images and yellow arrows indicate phantom-shaped bright regions aliased onto the real image:
 |
| Through plane reconstruction for 1.5 mm isotropic resolution scans, SMS=6: TL: SBref. TR: MB-EPI. BL: Blipped-CAIPI, 5x5 kernel, CF=1. BR: Blipped-CAIPI, 5x5 kernel, CF=2. (Click to enlarge.) |
Let's summarize things so far. The images from 2.5 mm and 2 mm protocols with SMS factors of 4 and 6, respectively, look good in qualitative terms. However, when the resolution is pushed to 1.5 mm, still with an SMS factor of 6, there are weak but visible reconstruction artifacts. For the Blipped-CAIPI sequence these artifacts are apparent whether compression factor 1 or 2 is used, suggesting that we can use CF=2 for reconstruction speed.
Effects of kernel size on image quality
I now want to go back a step and look at the kernel size. Above I selected the 5x5 kernel option throughout, while noting that the 3x3 and 3x5 options weren't as good. Here is a comparison of the 3x3 kernel to the 5x5 kernel using the 2 mm isotropic resolution protocol at SMS factor of 6. Top left is SBref, top right is MB-EPI, bottom left is Blipped-CAIPI with the 3x3 kernel and bottom right is Blipped-CAIPI with the 5x5 kernel:
 |
| Ghost contrasted 2 mm isotropic resolution scans, SMS=6: TL: SBref. TR: MB-EPI. BL: Blipped-CAIPI, 3x3 kernel, CF=1. BR: Blipped-CAIPI, 5x5 kernel, CF=1. (Click to enlarge.) |
The artifacts are quite similar between the MB-EPI sequence and Blipped-CAIPI reconstructed with the 3x3 kernel. Does this mean the CMRR recon uses a 3x3 kernel? I don't know but shall endeavor to find out. The 5x5 kernel gives the crisper N/2 ghosts that better resemble the SBref "ideal" ghosts.
I also ran some 50-volume time series acquisitions that showed reduced temporal signal-to-noise ratio (tSNR) for the 3x3 kernel option:
 |
| TSNR images for 50-volume time series measurements: 2 mm isotropic voxels, SMS factor 6. Left: MB-EPI. Middle: Blipped-CAIPI, 3x3 kernel, CF=1. Right: Blipped-CAIPI, 5x5 kernel, CF=1. |
The reduced tSNR comes from the spatial correlation of the artifacts across the entire image plane. Note the smoothness of the artifacts in the center panel and how they propagate across all the background regions:
 | |||
| Standard deviation images for 50-volume time series measurements: 2 mm isotropic voxels, SMS factor 6. Left: MB-EPI. Middle: Blipped-CAIPI, 3x3 kernel, CF=1. Right: Blipped-CAIPI, 5x5 kernel, CF=1. |
So we can conclude that the cost of the smaller kernel is higher artifacts, reduced temporal stability and lower power, which doesn't look like a good trade to me. I would prefer to use the 5x5 kernel and set the compression factor to two for speed, although at this point I haven't yet done a full temporal analysis to know that setting CF=2 wouldn't similarly degrade tSNR, of course.
Blipped-CAIPI performance on a brain
And so, finally, to the brain. I want to emphasize that whatever you see here should be taken with an even larger pinch of salt than the phantom tests! It's very difficult to discriminate artifacts with brain anatomy getting in the way. But this is our first opportunity to check for new problems, such as an artifact from exquisite motion sensitivity. I'm not actively testing motion sensitivity here, however. That has to wait for a later post.
For now I offer only 3D multi-planar views for the three starting point protocols at 2.5, 2.0 and 1.5 mm isotropic resolution using compression factors of 1 and 2 (but 5x5 kernel throughout), zoomed and contrasted independently to provide the most comprehensive view of the brain:
 |
| 2.5 mm isotropic resolution, Blipped-CAIPI sequence, SMS factor 4, 5x5 recon kernel, CF=1. |
 |
| 2.5 mm isotropic resolution, Blipped-CAIPI sequence, SMS factor 4, 5x5 recon kernel, CF=2. |
 |
| 2 mm isotropic resolution, Blipped-CAIPI sequence, SMS factor 6, 5x5 recon kernel, CF=1. |
 |
| 2 mm isotropic resolution, Blipped-CAIPI sequence, SMS factor 6, 5x5 recon kernel, CF=2. |
 |
| 1.5 mm isotropic resolution, Blipped-CAIPI sequence, SMS factor 6, 5x5 recon kernel, CF=1. |
 |
| 1.5 mm isotropic resolution, Blipped-CAIPI sequence, SMS factor 6, 5x5 recon kernel, CF=2. |
What can I conclude from these views? Only that I don't see any obvious new problems requiring further investigation. Together with the phantom tests I would set my personal limits at 1.5 mm isotropic resolution and SMS factor 6. I would only push to 1.5 mm resolution if the experiment absolutely necessitated it, however. I wouldn't want to use an SMS factor greater than 6 for any protocol without a lot more testing, and right now testing a factor of 8 is low on my list of priorities.
With these starting points and a basic evaluation of two SMS sequences in the bag I'll be moving on to more detailed investigations of the consequences of slice-to-slice cross-talk and motion sensitivity.
____________________
Notes:
1. From Setsompop et al. (2012):
"The simultaneous multi-slice method does put some constraints on the number of slices. For example, the acquisition is simplified if the total number of slices is a multiple of Rsl. A more subtle effect occurs when an interleaved slice order is used. The purpose of interleaving is, of course, to avoid exciting spatially adjacent slices in rapid succession. In a standard interleaved acquisition, adjacent slices are taken TR/2 apart in time. The interleaved Rsl = 3 acquisition has an additional constraint if one wishes to avoid having some spatially adjacent slices acquired in rapid succession. In the simultaneous multi-slice acquisition with a total of Nsl, a total of Rsl subgroups each with Nsl/Rsl slices are created. The successive excitation problem occurs between the top slice of one subgroup and the bottom slice of the subgroup above it. The problem can be avoided if the number of slices in each excitation subgroup is odd. Thus Nsl/Rsl should be an odd integer to avoid signal loss slices with imperfect slice profiles at the edge of each sub-group. If an even integer is chosen, the first slice of each subgroup will be excited right after the excitation of an adjacent slice that corresponds to the last excited slice in an adjacent slice group (from the previous TR). This leads to a signal loss from the slice crosstalk for these edge slices."
An illustration of the cross-talk is shown in Fig. 3 of Barth et al. (2016) review of SMS methods:
2. I'll do a post at a later date that looks at the issue of SMS support in relation to the RF coil being used. A colleague observed that there are only about five discrete rings of coil elements in the Z direction for the Siemens 32-channel coil. The implication seems to be that the blipped CAIPI scheme is doing a lot (most?) of the work of separating the simultaneous slices once the SMS factor gets above 5.

Hi,
ReplyDeleteI think it is worth explicitly mentioning here that it is inherently more difficult/challenging to unalias the data without artifact in a spherical homogeneous AGAR phantom than in a human. Which is to say that a set of parameters that appear borderline or even outright unreasonable in the AGAR phantom, may actually be fine to use in a human. As you know, the HCP is comfortable using MB=8 in humans using the CMRR sequences and recon, even though we can indeed see some recon artifacts at MB=8 in the AGAR phantom. :)
cheers,
-Mike Harms
Hi,
ReplyDeleteWere there any updates on the optimized sequences? Our lab is now testing MB-EPI and trying to find a good balance between all the parameters.
Also, could you please update the Dropbox link for the PDF with the complete sequence parameters?
Thank you!
We're basically using the CMRR sequence as listed in the Part I post. We have generally restricted the MB factor to between 2 and 5 for our 32ch coil. MB=3 seems to be a good compromise for many people. Spatial resolution is generally (2.5 mm)^3 regardless of MB factor. Higher resolution drives the echo spacing and TE to the point where partial Fourier is needed to keep the TE reasonable, whereas there's no need to use partial Fourier for 2.5 mm voxels. We have started to use Leak Block (aka split slice GRAPPA), but for MB < 5 and the 32ch coil the leakage is already pretty low, so the differences with/without are small.
DeleteHere's an example parameter set for MB=4:
https://www.dropbox.com/s/9jsx585c5kl6p5z/mb_bold_mb4_2p5mm_AP_PSN.pdf?dl=0
Here, LeakBlock is on, and we have Prescan Normalize enabled with the option to also save the raw images. Note that the total number of slices is odd, 60/4 = 15, per the suggestion in Barth et al (2016), in the above post.
One option you may want to investigate, especially if you're mainly interested in frontal lobe, is to reverse the phase encoding direction, using the Invert RO/PE Polarity option on the Special Card. Our frontal lobe researchers use this and like it.
LMK if I can answer more questions.